Publications Hub, Clinical & Translational Research, Article
Revisiting the subgingival microbiota: Towards an ecological risk scale for periodontology
09 September 2025
A greater understanding of the microbiota beneath the gumline could open the way to predicting the course of periodontal disease, further enhancing diagnostics beyond the current approach of staging and grading. Julien Santi-Rocca, winner of the first prize in the Jaccard-EFP Research Prize 2025, and his colleague David Felipe Martín-García explain the development and implications of their research into developing a microbial risk scale.
Disease staging and grading—the key innovation of the 2018 classification of periodontal and peri-implant diseases—has advanced periodontal diagnostics by measuring tissue damage and evaluating its progression. Yet, this approach relies on the measurement of clinical outcomes and risk factors (such as smoking or diabetes) that are beyond the dentist’s control.
What if we could diagnose the threat before any damage occurs? What if the microbial ecology beneath the gumline offered a predictive signal—not just a record of disease history but also a glimpse of its future trajectory?
Rather than focusing on identifying new pathogens, our team set out to understand the organization of the subgingival microbiota. Could this ecosystem be read as a structured system, with its own dynamics and states? Twenty-five years ago, Sigmund Socransky and his colleagues defined, among 40 pre-selected species, five bacterial complexes—an ecological framework that shaped our understanding of disease progression. Today, we believe it is time to build on this landmark contribution, using modern tools that allow us to unravel microbial interactions without prior assumptions.
Building a new microbial framework
Using high-resolution meta-taxonomic sequencing combined with a probabilistic taxonomic assignment method (PRONEX), we explored the bacterial landscape of the subgingival space across 135 samples. What emerged was not a simple health-to-disease gradient, but five distinct microbial profiles—ecological states—defined by consistent patterns of co-abundant bacterial taxa.
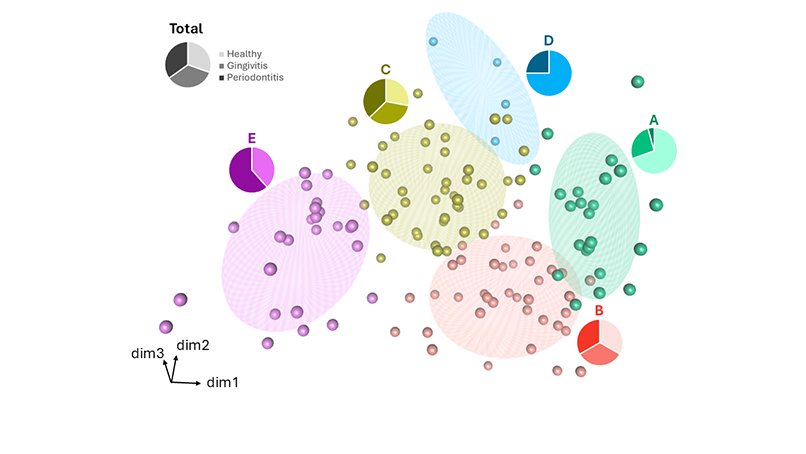
The mismatch between microbial patterns and clinical diagnoses could be explained by a temporal lag: ecological shifts may precede tissue damage. This opens the door to predictive or preventive diagnostics, where microbiota profiling helps identify patients at risk before any symptoms appear. Similarly, during treatment, microbiota shifts could help anticipate therapeutic outcomes and enable personalized strategies.
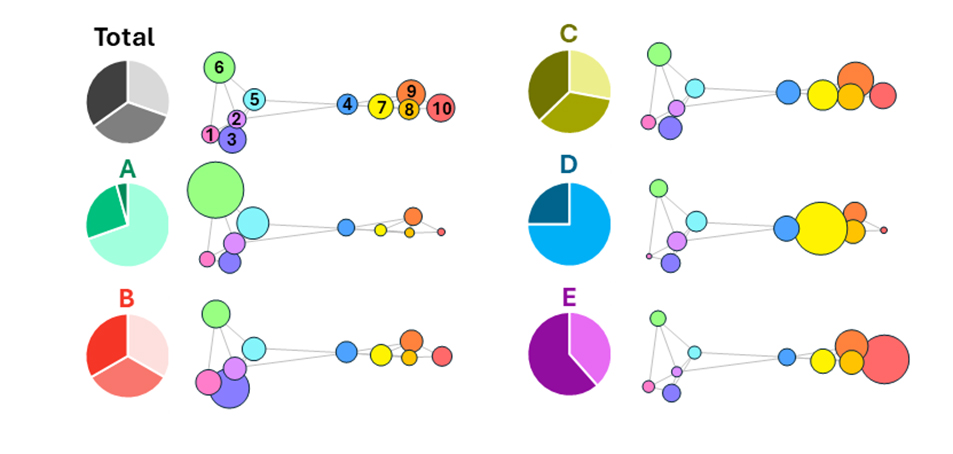
Microbial complexes: something old, something new
This study benefited from technological advances and allowed us to refine Socransky’s vision of microbial complexes. While his 32 targeted taxa represented 30.9% of the sequences obtained in our study, the 162 taxa we selected covered 95.0% of these sequences. The precision gained, as well as the number of analysed taxa, enabled the identification of 10 microbial complexes—clusters of co-abundant and co-occurring taxa. Some match Socransky’s original groups (e.g., Complex 10 comprises the species from the “Red Complex”), while others diverge or are newly described.
What matters is not only the composition of each group but also their combination: certain proportions align with clinical health, others with periodontitis, and some with the previously defined microbial patterns. These complexes can be considered fundamental ecological units of the subgingival biofilm. Their relative abundance may define the commensal or pathogenic nature of the ecosystem.
Furthermore, some taxa are tightly associated with a single complex and may serve as core markers; others act as boundary taxa and could signal ecological transitions.
The microbial risk scale: a new diagnostic dimension
This framework opens the door to a third diagnostic axis: microbial risk. This axis can complement staging and grading by positioning the patient's microbiota within a dynamic ecological landscape:
- Are there consistent markers of health or disease? → Complexes.
- Is the ecosystem stable or tipping? → Boundary taxa.
- Does the profile follow a typical disease path or indicate an alternative route? → Patterns.
These questions reframe diagnosis as anticipatory—using microbiota profiling to estimate risk before irreversible damage occurs. Notably, one microbial pattern (Pattern D) was associated with disease but lacked P. gingivalis, the classical keystone pathogen, suggesting the existence of alternative disease trajectories that may involve other microbial kingdoms or provide evidence of host specificities.
From sequencing to practice: making the leap
Sequencing is unlikely to become a routine diagnostic tool in the immediate future, but innovation is narrowing the gap. Advances in multiplex quantitative polymerase chain reaction (qPCR), lab-on-chip systems, and AI-assisted platforms are making molecular microbiota profiling more accessible. These tools may soon support risk stratification, monitoring, and personalized interventions in periodontal care.
In the meantime, our findings foster the use of phase-contrast microscopy as a powerful and underused diagnostic surrogate. With minimal equipment, clinicians can observe morphotypes and motility—key indicators of the ecosystem’s state. For instance, Complex 10, linked to periodontitis, includes nine Treponema species, easily recognised as motile and spiralled bacteria under the microscope. This contrasts with the sessile morphologies observed in health-associated complexes.
Future directions: towards ecological and precision medicine
These insights contribute to the emergence of predictive, ecological medicine—where diagnosis is no longer just retrospective but also guides early, personalized prevention. These tools can be used for the early follow-up of therapy because shifts in the microbiota might precede healing. We are currently working on:
- Refining the patterns by analysing the non-bacterial microbiota in the samples: parasites, fungi, archaea, and viruses.
- Building innovative diagnosis tools for immediate, molecular-based multiplexed biomarker analysis of periodontal risk to use at the clinic.
- Developing image-recognition algorithms for microscope-based risk estimation.
Our results can also guide microbiota-based interventions beyond diagnosis:
- Seeding with core taxa associated with health as controlled microbiota grafts for preventive intervention or as adjuvants during periodontal treatment.
- Targeting core taxa associated with disease to limit damage.
- Controlling boundary taxa to prevent the evolution towards disease.
Above all, we aim to codevelop these tools with clinicians because diagnostic and therapeutic tools only change practice when they are trusted, used, and compatible with real-world care.
We invite clinicians, researchers, and partners to join this effort. Together, we can move beyond damage assessment and towards the early detection of imbalance, where the microbiota becomes not just a trace of past disease but also a guide to preserving health.
Biography

Julien Santi-Rocca, PhD, and David Felipe Martín-García, PhD, are the founders of SHOW (Science and Healthcare for Oral Welfare), a research initiative focused on microbial ecology and translational diagnostics and therapeutics. Julien, recipient of the 2025 Jaccard-EFP Research Prize, leads the biological research, while David brings expertise in bioinformatics and societal impact, helping to ensure that innovations remain accessible and aligned with clinical practice. Together, they work to bring data-driven, ecologically grounded insights from bench to chairside in periodontology.
See also:
Journal of Clinical Periodontology: Julien Santi-Rocca, David F. Martín-García, Iván Lorca-Alonso, et al. "Microbial Complexes in Subgingival Plaque: A Bacterial Meta-Taxonomic Study". Journal of Clinical Periodontology 52(7), July 2025, 983-998. https://doi.org/10.1111/jcpe.14138
News report: 'Jaccard-EFP Prize awarded at EuroPerio11'