Publications Hub, Clinical & Translational Research, Article
How shotgun metagenomics can improve early diagnosis of peri-implant diseases
04 September 2025
Peri-implantitis affects about one in five implant patients and accurate early diagnosis remains a significant clinical challenge. Paolo Ghensi reports on the work he and his research team at the University of Trento (Italy) are doing to see how microbiome analysis might improve the situation. Their research recently received the third prize in the Jaccard-EFP Research Prize 2025.
Current diagnostic methods for peri-implantitis—primarily based on bleeding on probing (BoP), suppuration, and probing pocket depth (PPD)—often detect disease only after substantial tissue damage has already occurred.
The limitations of these traditional diagnostics are well known to clinicians. While BoP and PPD measurements are essential clinical tools, their sensitivity to early disease detection is modest at best. By the time these parameters become clearly positive, the disease process is often well established. This diagnostic gap has real consequences—both clinical and economic—as late detection frequently necessitates complex (and costly) surgical interventions.
Microbiome analysis offers new diagnostic possibilities
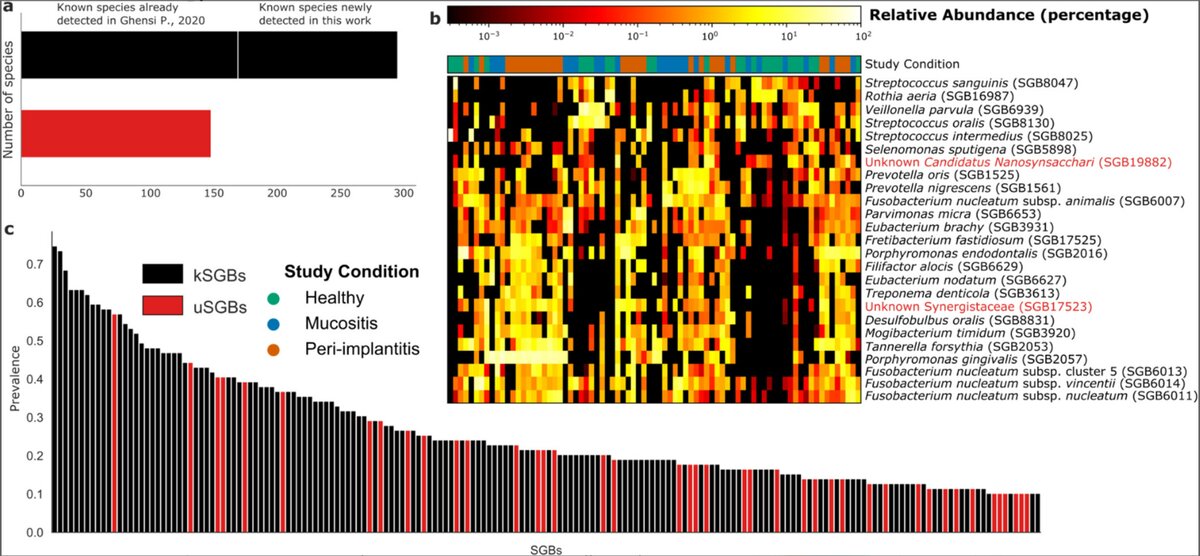
Our cross-sectional study applied shotgun metagenomics technology to analyse subgingival plaque microbiome samples from 158 sites across 102 patients, comparing healthy implants, mucositis, and peri-implantitis cases. Using MetaPhlAn 4, a computational tool that can identify microorganisms at the species and strain level, we catalogued the complete microbial communities present at each site.
The results revealed 447 different bacterial species in the peri-implant environment—considerably more diversity than had been suggested in previous studies using traditional methods. Perhaps more surprisingly, 150 of these species (34%) were previously uncharacterized microorganisms that current databases cannot yet name. Despite being "unknown”, many of these species showed strong associations with specific disease states.
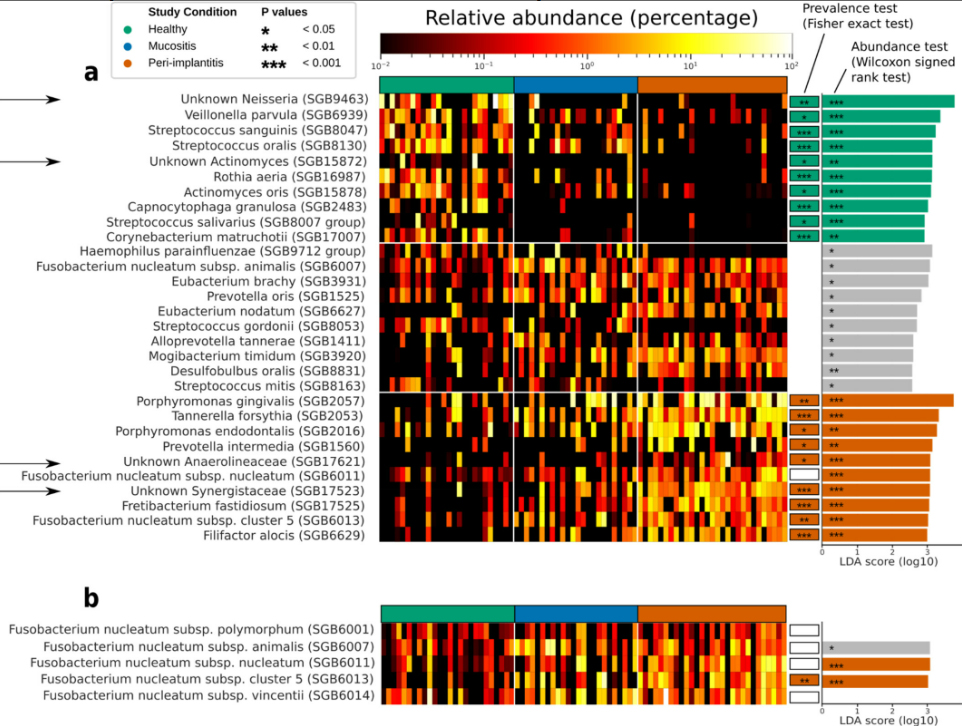
Each clinical condition displayed its own characteristic microbiome pattern. Healthy sites maintained diverse microbial communities that varied considerably between individuals. In contrast, peri-implantitis sites showed much more consistent microbiome profiles across different patients, suggesting that disease drives convergence toward specific pathogenic communities. Mucositis appeared as an intermediate state, sharing features with both healthy sites and disease sites.
Machine learning enhances diagnostic precision
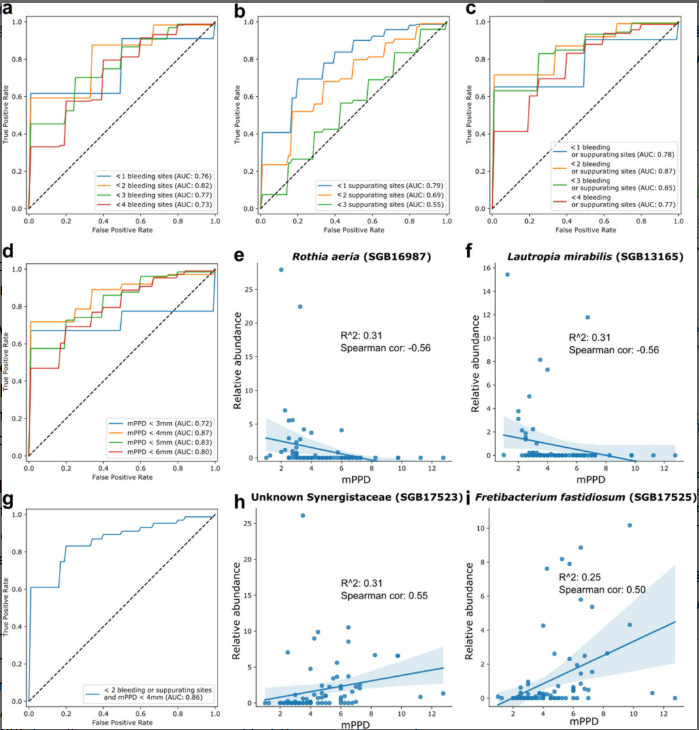
To translate these complex microbiome profiles into clinically useful information, we employed machine-learning algorithms—specifically random-forest classifiers—to identify which microbial patterns best predicted each clinical condition. The accuracy of these models was good to excellent, with area-under-the-curve (AUC) values between 0.78 and 0.96 depending on the specific comparison.
These accuracy levels are encouraging, particularly for distinguishing peri-implantitis from healthy sites (AUC 0.96). The models also showed meaningful correlations with traditional clinical parameters, validating the concept that the microbial signatures reflect genuine biological processes rather than statistical artifacts.
One particularly interesting finding involved Fusobacterium nucleatum, a species long associated with periodontal disease. Our high-resolution analysis revealed that different subspecies of F. nucleatum play distinct roles—some associated with health, others with mucositis, and with specific strains strongly linked to peri-implantitis. This subspecies-level discrimination would be impossible with conventional diagnostic methods.
Practical applications in clinical settings
How might this technology translate to everyday practice? The most straightforward application would be risk assessment for implant patients. During routine maintenance visits, microbiome analysis could identify patients who are developing pathogenic communities before clinical signs appear. This early warning system could trigger preventive interventions—such as more frequent professional debridement, targeted antimicrobial therapy, or closer monitoring.
Treatment monitoring represents another promising application. Rather than relying solely on clinical parameters to assess treatment response, microbiome analysis could provide objective evidence of whether therapeutic interventions are successfully shifting the microbial community back toward health. This could help clinicians make more informed decisions about treatment duration and intensity.
The sampling process itself is straightforward, similar to current microbiological sampling methods; samples can be collected during routine examinations using sterile curettes, although proper handling protocols are essential for obtaining reliable results.
Comparing approaches: shotgun metagenomics versus 16S sequencing
Many clinicians are familiar with 16S rRNA sequencing, which has been the standard molecular approach for microbiome studies. While 16S analysis is valuable, shotgun metagenomics offers several advantages for clinical applications. Most importantly, it provides species and strain-level identification rather than just genus-level information. This matters because, as our F. nucleatum findings demonstrate, different strains of the same species can have opposing roles in health and disease.
Shotgun sequencing also captures the entire microbial community—bacteria, viruses, fungi—and their functional genes. This comprehensive view enables detection of antibiotic resistance genes and virulence factors that could inform treatment selection.
Challenges and considerations for implementation
Despite promising results, several hurdles remain before this technology becomes routine in dental practice. Cost is a key consideration because shotgun sequencing is more expensive than 16S sequencing—although prices are declining sharply as the technology improves. However, if early detection prevents even a fraction of complex peri-implantitis cases, the economic argument in favour of shotgun sequencing becomes more favourable.
Standardization presents another challenge. Sampling protocols and analysis pipelines must be rigorously standardized to ensure reproducibility across different settings. This will require coordinated efforts between research groups and professional organizations.
Interpretation of results also requires careful consideration. While our models showed strong predictive accuracy in the research setting, real-world performance may vary. Clinicians will need training to understand and act appropriately on microbiome reports, integrating this new information with traditional clinical diagnosis.
Looking ahead
This research suggests that microbiome-based diagnostics could meaningfully improve peri-implant disease management, but we should maintain realistic expectations. This technology will not replace clinical examination but rather will complement it, providing additional information to guide treatment decisions.
The discovery that many important biomarker species remain uncharacterized highlights just how much we still have to learn about the peri-implant microbiome. Future research should focus on understanding these unknown organisms’ roles in disease processes, potentially revealing new therapeutic targets.
For now, clinicians should stay informed about these developments while continuing to rely on established diagnostic and treatment protocols. As validation studies progress and costs decrease, microbiome analysis will likely find its place in the diagnostic toolkit, gradually expanding to the routine care of everyday practice.
The ultimate goal remains unchanged: detecting and treating peri-implant diseases early enough to preserve implant health and function. If microbiome analysis can contribute to this goal—as our research suggests it might—then the effort to translate this technology into practice will be worthwhile.
Select bibliography
Berglundh T, Armitage G, Araujo MG, et al. Peri-implant diseases and conditions: Consensus report of workgroup 4 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J Clin Periodontol 2018;45: S286-S291.
Blanco-Míguez A, Beghini F, Cumbo F, et al. Extending and improving metagenomic taxonomic profiling with uncharacterized species using MetaPhlAn 4. Nat Biotechnol 2023;41: 1633-1644.
Caton JG, Armitage G, Berglundh T, et al. A new classification scheme for periodontal and peri-implant diseases and conditions: Introduction and key changes from the 1999 classification. J Periodontol 2018;89: S1-S8.
Derks J, Tomasi C. Peri-implant health and disease. A systematic review of current epidemiology. J Clin Periodontol 2015;42: S158-S171.
Heitz-Mayfield LJA, Salvi GE. Peri-implant mucositis. J Periodontol 2018;89 Suppl 1: S257-S266
Ghensi P, Manghi P, Zolfo M, et al. Strong oral plaque microbiome signatures for dental implant diseases identified by strain-resolution metagenomics. npj Biofilms Microbiomes 2020;6: 47.
Pasolli E, Truong DT, Malik F, Waldron L, Segata N. Machine learning meta-analysis of large metagenomic datasets: Tools and biological insights. PLoS Comput Biol 2016;12: e1004977.
Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol 2017;35: 833-844.
Schwarz F, Derks J, Monje A. Peri-implantitis. J Clin Periodontol 2018;45: S246-S266.
Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. Microbial complexes in subgingival plaque. J Clin Periodontol 1998;25: 134-144.
Tett A, Pasolli E, Farina S, et al. Unexplored diversity and strain-level structure of the skin microbiome associated with psoriasis. npj Biofilms Microbiomes 2017; 3:14.
Thomas AM, Manghi P, Asnicar F, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat Med 2019;25: 667-678.
Biography

Paolo Ghensi (DDS, Oral Surg. MClin Dent, Clin MSc, PhD) is a clinician and researcher specializing in periodontology, implantology, and oral surgery. He holds a PhD in biomolecular sciences and has received recognition for his research contributions, including the SIdP Henry M. Goldman Prize (2018) for basic research. An active member of the IAO and SIdP with SIdP board certification in periodontology and implantology, he has authored over 50 scientific publications focusing on peri-implant diseases, oral-microbiome studies, and regenerative dental procedures. His clinical expertise includes periodontal therapy, computer-guided implantology, bone-regeneration techniques, and soft-tissue management around teeth and implants.
See also:
Journal of Clinical Periodontology: Paolo Ghensi, Vitor Heidrich, Davide Bazzani, et al. 'Shotgun metagenomics identifies in a cross-sectional setting improved plaque microbiome biomarkers for peri-implant diseases'. Journal of Clinical Periodontology 52(7), July 2025, 999-1010. https://doi.org/10.1111/jcpe.14121
News report: 'Jaccard-EFP Prize awarded at EuroPerio11'




